Sunday, July 10, 2011
Sunday Spinelessness - The first animals (molecular biology)
It's time to wrap up this series of posts on the origin of animals. If you are just tuning in here, I've already decided that early fossils are utterly fascinating but really not much help to us and had a look at some modern organisms that might give us an idea about how the major steps towards multi-cellularity might have been achieved. Today, I'm going to zoom down another level, and see if molecular biology can tell us anything about the first animals.
Life is really just a very special kind of chemistry. The organisms that emerge from the bewildering number of inter-connected chemical processes that make life tick are so complex, so varied and so far removed from the things that chemists study that we had to create a whole other science, biology, to study them. For a long time biologists and chemists more or less got on with their own problems, but in the middle of the 20th century the progress each field had made created the opportunity to actually study the molecules that make life. The so called "molecular revolution" in biology has effected almost every sub-discipline within our science, and the tools and data that revolution created can be used to try and understand the origin of animals. Over the last decade or so scientists have sequenced the genomes of an increasingly number of species, and in between the big headlines created by creatures like chimps, mice and humans; we've been able to learn about sponges, cnidarians and placozoans. Those sequences can help us understand how these animals work at a molecular level, and they can also be used to recover the relationships between different groups of animals.
Scientists call the branching trees we create to represent the relationships between species 'phylogenies', and as someone who spends quite a lot of his time working on molecular phylogenies (have you seen The Atavism's logo...) I should really be saying that this is the best way to understand the origin of animals. But that's not really true. DNA isn't quite the wonder-molecule that CSI may have lead you to believe it is, and having DNA sequences available isn't necessarily enough for us to recreate a phylogeny. At the moment, the relationships at the base of the animal family tree are very hard to untangle. We know that a group of animals called the bilatarians (you, me, insects, fish, sea urchins, nematodes, spiders and, well, really every animal you are likely to name off the top of your head) form one group distinct from the other animals, and that these animals can be further subdivided based on the way their embryology plays out. But the relationships of the non-bilatarian animals - a grab-bag of creatures including the cindarians (jellyfish, corals and their relatives), comb jellies, placozoans and sponges - are really unclear:
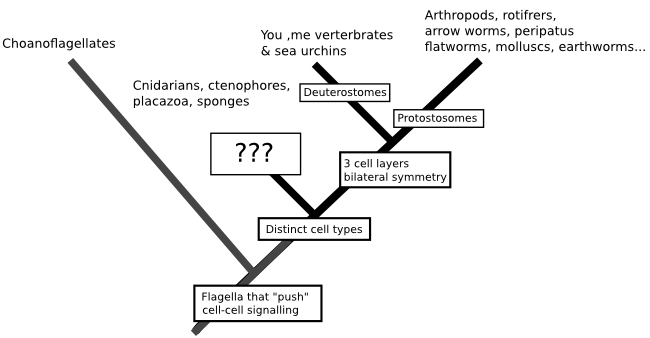
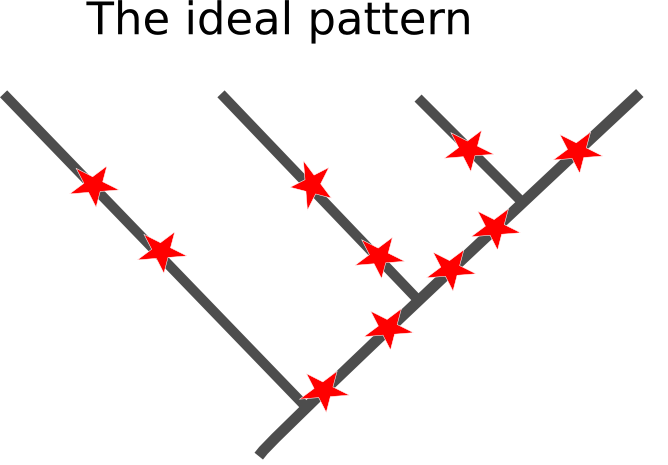
That uncertainty isn't a matter of us just not having enough data to throw at the problem. Molecular phylogeny works by grouping species together based on mutations that they share, having inherited them from a common ancestor. So, ideally all the branches in the true history of the species we are studying will be separated by enough time for those mutations to start racking up in the period of time that lineages shared an evolutionary history, something like this (with the mutations being the red stars occurring at random):
Sadly, the real pattern underling the base of the animal family tree doesn't appear to be like this. The initial branches fromed relatively quickly, leaving only short periods of shared ancestry for related groups to accrue mutations that would allow us to recreate their relationship. Worse yet, the branches that lead out from initial splits are really long, which means there has been millions of years for any useful mutations to have been over-written by more recent changes in DNA sequences.
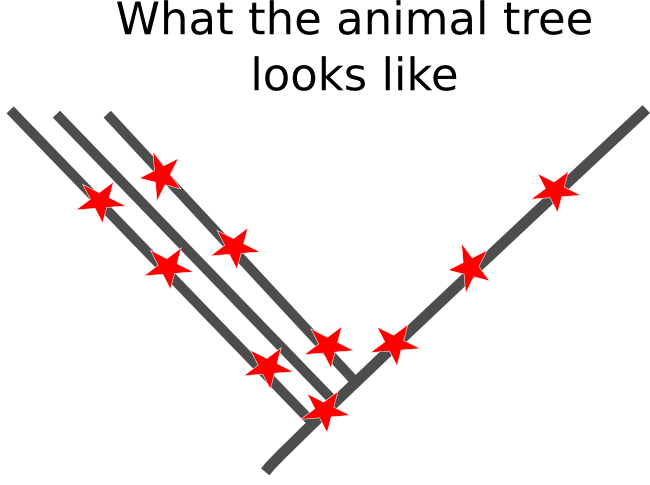
There is no easy way to get around this problem. It seems like we should be able to throw more and more genetic sequences at the tree, increasing the chance the we get some informative mutations, but these trees run the risk of falling into the terrifying Felsenstein zone, in which the confidence with which we estimate the wrong tree increases as we apply more sequences to the problem. Because of the difficulties inherent in reconstructing the very early history of animals, I think we should take results based solely on molecular phylogenies with more than a few grains of salt.
A case in point is the phylogeny Casey Dunn and colleagues presented in Nature a few years ago (doi: 10.1038/nature06614). If we were trying to fill in the question marks and the base of the animal tree without considering molecular data at all, we'd probably put sponges on the first branch to split in the animal family tree. Sponges are quite different from other animals, they don't have a nervous system, their cells are not bound into tissues and, although sponges do have specialised cell-types, their cells are uniquely flexible in that mature cells can change from one type to another. It's actually quite possible that several of the early branches in the animal tree lead to different types of sponges, like this:
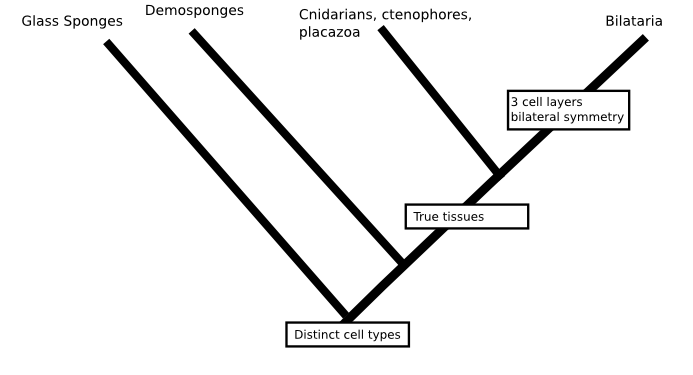 But, against all expectations, Dunn's phylogeny found comb jellies (creatures that look a little like jelly fish but form a phylum, Ctenophora, in their own right) to be the first group to split from the animal tree. If that were true, then we'd be less confident that the first animals were sponge-like since either (a) sponges would be radically modified animals which had lost their nervous system, given up on tissues and taken to a sedentary filter feeding life or (b) comb jellies would have evolved nervous systems and tissues independent of those of other animals.
But, against all expectations, Dunn's phylogeny found comb jellies (creatures that look a little like jelly fish but form a phylum, Ctenophora, in their own right) to be the first group to split from the animal tree. If that were true, then we'd be less confident that the first animals were sponge-like since either (a) sponges would be radically modified animals which had lost their nervous system, given up on tissues and taken to a sedentary filter feeding life or (b) comb jellies would have evolved nervous systems and tissues independent of those of other animals.
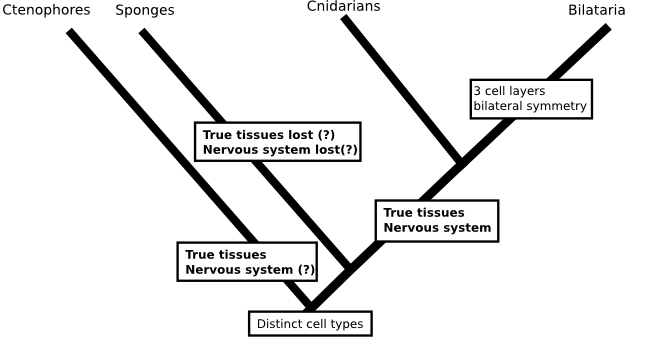
There is no reason that such a scenario would be impossible (in fact, fungi and plants have each evolved tissues independently of animals), but when the whole idea is constructed from a method that we know is prone to biases I don't think there is much point in worrying about it (I should say, this isn't a criticism of the paper I'm talking about, which took all the sensible measures to deal with the problems inherent in there analysis and presented their tree as something to work on rather than a final result). Similarly, a phylogeny that included data from everyone's favourite animals the placozoans (Schierwater et al., 2009. doi: 10.1371/journal.pbio.1000020) was used to resurrect a hypothesis for the origin of animals that includes a flat ancestor, not unlike a modern placazoan or the 'planula' larvae of some cindarians. This scenario would again require nervous systems to evolve twice:
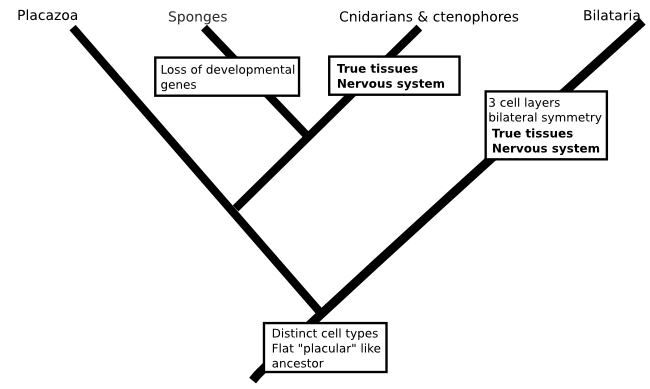
One of the reasons I'm skeptical of this idea comes from what we've learned about the genomes of simple animals. For my money, the discovery of Hox genes is the most remarkable result to come out of the the molecular revolution I talked about in the intro to this post. In turns out the genes that orchestrated your embryonic development have counterparts in Drosophila (or Sophophora if you'd rather) that do exactly the same job. If we move away from the bilataria, Hox genes are a bit harder to come by, but they still exist in the genomes of comb jellies, cnidarians and even palcozoans. But sponges have no Hox genes (Larroux et al, 2007. doi: 10.1016/j.cub.2007.03.008 and references therein). It's conceivable that the sponges descend from an ancestor that had Hox genes, then gave them up, but it seems much more likely that Hox genes evolved after sponges parted ways with the rest of the animal kingdom.
The contents of animal genomes tell us more than which phylogenetic patterns are likely - they can also let us know how some of the unique features of animals might have arisen. Almost every time the genome sequence of a simple animal is published you'll get a press release saying how surprising it is that genes associated with functions of more complex animals were found in this simple creature. And every time I read one of these press releases I sigh. Thesy are exactly analogous to someone that studies birds expressing surprise that therapod dinosaurs (the ancestors of birds) had forelimbs even though they couldn't fly. You can't make entirely new gene families out of nothing, new functions evolve when existing genes are retooled to attack different problems. The first animals faced two big problems when they moved from a single-celled lifestyle to a mulit-celled one: first, they needed to hold themselves together; and second, they needed ways for cells to communicate to their neighbours. Modern animals stick together with collagen (an entirely animalian invention) and a bunch of other sticky proteins including cadherins. The closest relatives of animals, the choanoflagellates which I talked about last week, have cadherins and other sticky proteins which they probably use to trap their prey (bacteria and other small cells).
Choanoflagellate genomes also give us a great insight into how the cells in the first animals started talking to each other. One of the most important pathways for cell-cell signalling in animals is called notch. Choanoflagellates don't have notch genes, but they do have the raw material from which one could be made. The proteins that notch genes make have several distinct 'domains' each of which perform particular function - and all these domains are present in choanoflagellate genomes! These different domains could have been brought together by genetic recombination, creating new genes by so called 'domain shuffling'. If we move just a little bit past the origin of animals and look at the genomes of sponges we can find the precursors of nervous system genes (see for example Liebeskinda et al., 2011. doi: 10.1073/pnas.1106363108).
So, that's three posts about 5 000 words and a whole bunch of figures on the origin of animals. What can we say we've learned? Certainly, studying modern animals seems to be the only way to approach the question of where animals came from. The models we looked at last week allow us to understand how the problems associated with a switch from single celled life, to a colonial lifestyle and finally the emergence of an individual identity for the colony. Modern genomes also tell us how those changes might have happened at the molecular level. Sometimes, as is the case of the cadherins and the evolution of the nervous system, existing genes could be re-purposed to a new tasks. Other times, new features were likely achieved with the help of new genes, cobbled together by genetic recombination. We can probably disregard theories for the origin of animals that require complex morphological changes based entirely on molecular phylogenies, since those estimates are prone to biases and the most interesting branches in them remain uncertain. If we look at morphological similarities between choanoflagellates and sponges (which goes right down to the collared and flagellated cells they use to eat) it seems likely that the first animals were filter feeders that pumped water into their cells with long flagella.
If you forced to make a bet on what the first animals were like, I reckon they'd be a bit like a sponge larva. A collection cells similar to a choanoflagellate colony, moving about in the water column with their flagella. The big question is how that individual could have emerged from the colony, and here I like Paul Rainey's ideas (described last week) about the way inter-cellular competition, which is bound to arise in a colony, could actually create the conditions that would foster cooperation. I'd be really interested to hear what any readers who made it all the way through this series think now.
Labels: animals, first animals, phylogenetics, sci-blogs, science, sunday spinelessness
4 Comments:
Really an unrelated and silly question comes to mind. I've been somewhat put off lately by claims made by physicists, astronomers, and planetary scientists (almost never bonafide biologists) that multicellularity is an inevitable result in the evolution of life on ANY planet capable of producing life. I'm of the opinion (probably because I'm a microbiologist, and am somewhat biased) that we've always been in the Age of Bacteria, that there have been so many fortuitous steps in our evolutionary history, esp. the early ones like the multiple endosymbiotic events that lead to the first eukaryotes, that to make the leap that multicellularity, animal and plant evolution seems like a stretch that no reputable scientist should be making. What's your take on this question?
I agree entirely, I always wince when I hear scientists talking as if intelligence was an inevitable result of evolution, as in "if there is life, there will eventually be intelligent life". Like you, I'd imagine the vast majority of planets that have life (if there are any others at all) probably have unicellular life (like earth did for most of its history).
And even if, in the long run, the factors that contribute to the evolution of multi-cellulariry are common, the chance of multi-cellular life becoming intelligent are pretty close to nothing! (I still like SETI though, it's worth a shot!)
Thanks for the comment, I should preface my reply by saying I really work at the other extreme of phylogeny - using gene trees in species delimitation etc, so probably don't read as deeply with regards problems at the other end.
I think morphology could help us in these questions, but I really don't know how well we model morphological change in phylogenetics (it feels as if it was left behind with parsimony). Perhaps it's more helpful in assessing the plausibility of molecular phylogenies by mapping traits on to them (but the risk here is putting such a strong prior on relatively simple trees w.r.t morphological data we can never overturn in with other data).
I don't if people are working on it, but once these genomes are really annotated it would be possible to use structural infromation from the DNA (presence or absence of protein folds, or protein families say) which might increase the resolution.




