Friday, May 1, 2009
weasel.py
So, the weasel script that I wrote to mock Dembski et al was kind of ugly, this is largely because I'm a terrible programmer but even I could improve on bits of it, and since I also thought I should learn about classes and dealing with command line arguments I spent some of a rainy weekend on a version that will collect paramaters from the commandline and make one run of the weasel algorithm with those paramters
Usage: weasel.py -[options] [popsize] [mutation rate]Makes one run of Dawkins' weasel algorithm, displaying the best string in each generation and the 'fitness' of that string (number of characters matching the target string). Options -h or --help display this help message -t target sequence (defaults to"Methinks it is like a weasel") -l use locking (once a character matchs the target do not mutate) -o [file] log the fitness of the each generation to the specified file Arguments popsize: the number of strings created in each generation mutation rate: the probability of each letter mutating in each geneartion (a floating point number between 0 and 1) Example python weasel.py -l -o outfile.csv 100 0.05 (a run with locking, logged to 'outfile.csv', n=100 and u=0.05)
Thursday, March 26, 2009
One last bit of weasling...
So, this weasel thing is actually quite a nice little toy for showing a few truths about how selection works in real populations. The latest complaint from the people that seek to bury evolutionary biology by attacking this toy is that it's not really an example of "cumulative selection" because it's possible for some letters that are already matching the target sequence to revert to a non-matching letter. If a letter can go 'backwards' then how can we say selection on the string is cumulative? Because in most populations most of the time most selection is "purifying"selection, holding on to the advances that have been made in past generations. It's easy enough to use the functions defined in the first post to simulate what would happen if selection stopped half-way through a run:
Removing selection pressure rapidly destroys strings

By choosing a random string in each generation after the 100th we soon loose all the ground we made. We also now that selection is a strong force in large populations and a relatively weak one in small populations.(The large difference in the variance in the n=1000 and n=100 treatments in yesterday's post is a result of this) So lowering the population size to 10 after 100 generations should weaken (but not destroy) the power of purifying selection:
Lowering population size weakens the power of selection

Which is what we see, even in this situation where selection is very weak and strings can actually loose ground in their road to the target string each new generation retains the effects of past generations positive selection (to get the fitness high int he first place) and purifying selection (to retain much more of that fitness in the face of mutation that we could expect by chance). So, each new generation starts with benefits of previous rounds of selection, selection is cumulative.
Wednesday, March 25, 2009
On the effects of locking weasels [updated]
For those of you new to the discussion the bright-lights of the ID movement seem to think Richard Dawkins' toy example of the power of random variation and selection, a program that 'evolves' Hamlet's mocking line to Polonius "Methinks it is like a weasel", is a bit of a cheat because it fixes correct letters in place once they match the target string. This is not true. If you want to follow the whole story Ian Musgrave is your man.
Just for fun I thought I look at how, and when, the 'locking' approach than the IDers think Dawkins used gets you the target phrase more quickly. Below is the result of 100 runs of the weasel program with and without locking and with various parameters
Effect of 'locking' on number of generations required to evolve target phrase

What you see above is the net result of pairs of "unlocked" and "locked" runs at different population sizes (n) and mutation rates (u). When the population in each generation is 100 and the chance of each character mutating is 0.01 there is a small (but significant) difference in number of generations required to get to the target. Up the mutation rate to 0.1 and the locking method comes into its own, with correct bases protected from the threat of deleterious mutations (which become much more likely as the "best" sting approaches the target) and mutations are effectively targeted to areas in which they are needed. Locked and unlocked programs are no longer significantly different when the population size is brought up to 1000 (since one of those 1000 strings is likely to have a beneficial mutation and retain all it's correct letters).
So, when the mutation rate is high 'locking' greatly speeds up the evolution of the target string by protecting the characters that are already matching and when population size is high it doesn't make much difference because both approaches will likely find a 'fitter' string that retains the matching characters of the parent.
So, at what mutation rates is the differences between the locking and non-locking approaches most stark, it's easy enough to run the weasel program at a bunch of different mutation rates:
The effect of Mutation Rate on generations required for locking and non-locking programs to converge on target string
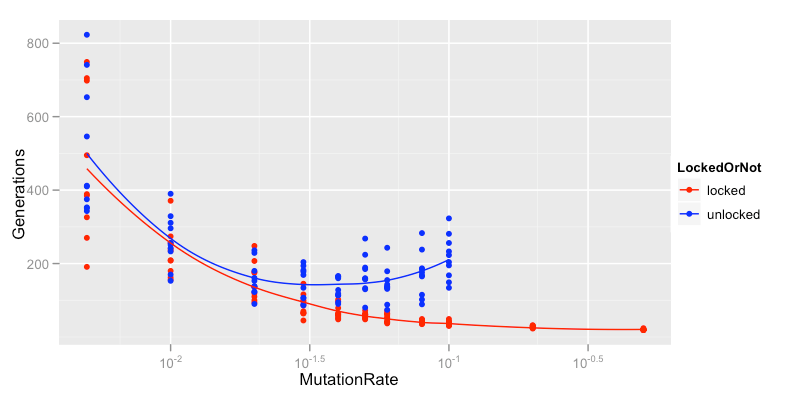
All these runs have a population size of 100. As you can see the locking approach is always quicker than the unlocked one but while the mutation rate is less than about 0.03 (3/popsize in this case) they are very similar. Once you raise the mutation rate above about 0.05 the unlocked mechanism actually gets slower as matching bases go unprotected against the the onslaught of mutation. This effect becomes so strong that for mutation rates above 0.1 the the algorithm can actually lose ground in its search for the target string:
Fitness of "best match" in one run of the weasel algorithm n = 100 u = 0.2

So, what have we learned from all this? The cumulative selection displayed in the "unlocked" weasel algorithm can certainly generate sentences that "blind chance" wouldn't arrive at during the lifetime of the universe. The 'locked' version of of the program that Dembski et al think Dawkins used is faster than the unlocked one but difference is trivial with relatively large population sizes and small mutation rates. All stuff that you could probably have worked out in first principles and took a dilettante programmer a few hours of spare time to show. Oh, and that the Evolutionary Informatics Lab still hasn't actually, you know, tried to test the things Dembski's been saying...
Labels: creationism, Dawkins, mutation, python, sci-blogs, weasel
Monday, March 23, 2009
Another Weasel
I know picking on creationists isn't clever or grown-up and posting notes to mock people so confused as to believe that the bible is a biology text book to a blog that has been left vacant for years borders on churlishness but this is just too cool not to contribute to.
As you may creationists having transitioned through cdesign proponentsism have recently evolved changed into Intelligent Design theorists. The unsophisticated of the past "it's all just too complex to have happened without god" have been replaced with sciency-sounding objections like "you can't get that much information by chance" or "you can't get that much complexity by chance". There are even whole labs devoted to proving this headed up by people with PhDs in maths!
It seems the leading lights of this new movement have spend the last several years worrying about a computer program that Richard Dawkins wrote on a a first generation Apple Mac in the 1980s. Dawkins' program is a toy aimed at showing just how powerful selection is as a force when compared to the blind "chance" that creationists often talk about. Dawkins showed that a room full of monkeys bashing keyboards (or a computer randomly choosing letters) would not merely fail to write Shakespeare's complete works in the lifetime of the universe - they'd be hard pressed to hit upon Hamlet's description of a cloud - "Methinks it is like a weasel". On the other hand, altering letters to a random sequence then selecting those most like a target to serve as the template for more rounds of variation and section can get you the phrase in a few minutes.
Ian Musgrave can tell you just how confused the creationists are by this program and provide details of their latest bizarre ideas. But the most interesting bit of all of this was the last line on a post from William Dembski the intellectual leader of the IDers:
In any case, our chief programmer at the Evolutionary Informatics Lab (www.evoinfo.org) is expanding our WEASEL WARE software to model both these possibilities. Stay tuned.
So the ID movement, which is a real and viable scientific objection to mainstream science and should be taught in higshchool and has "information science" labs and everything is working on recreating a program than an ethologist created in BASIC in the 1980s! Just for fun I wrote my own weasel program in python this morning (I was away for the weekend so only caught up with this on my return) which I share with you below. Keep in mind here I am not a real programmer, i have learnt to write a few scripts in BioPython for a research project but my skills are really very lacking. If the "chief programmer at the Evolutionary Informatics Lab" can't do this then what are they doing?
(You should also check out this completely different Pythonic weasel program from Anders Gorm)
*edited to intigrate the locked/unlocked bit Dembski et al have been talking about as an optional argument to make_gen() and add a bit to the comments
# -*- coding: utf-8 -*-
# Dawkins' weasel algorithm from /The Blind Watchmaker/ wrought in python
import string
from random import Random
def make_initial(target):
'''Make a random string, containing upper and lower case letters
and the same length as the target sequence for the run
'''
return ''.join(Random().sample(string.letters + ' ', len(target)))
def mutate(template, mutationrate):
'''Mutate a string at given rate without locking in chars that match the
target. The arguments are the template string followed by the probability
that each char is changed (there is a ~1/50 mutated character will actually
remain the same since the replacement is selected from the same pool of
characters)
'''
l = list(template)
for i in range(len(l)):
if Random().random() <= mutationrate:
l[i] = Random().sample(string.letters + " ", 1)[0]
else:
continue
return ''.join(l)
def mutate_locked(template, mutationrate, target):
'''Mutate a string but if any any chars already match the target sequence
leave them alone. Arguments are the template string, probability
that each char is changed and the target string
'''
l = list(template)
for i in range(len(l)):
if l[i] == target[i]:
continue
elif Random().random() <= mutationrate:
l[i] = Random().sample(string.letters + " ", 1)[0]
else:
continue
return ''.join(l)
def make_gen(template, target, popsize, mutationrate, locked = False):
'''Makes a new generation from a template string by adding new
mutants (from mutate() above) then scores each against the target string
and returns WeaselGen instance (defined above)
The arguments are the template string to base the new genetation, the
target string (to score offspring), the size of the population to make,
the rate of mutation at each char and whether or not characters are
locked in place once they match the target (a boolean defaulted to
false)
'''
generation = []
genscores = []
if locked == True:
for i in range(int(popsize)):
generation.append(mutate_locked(template, mutationrate, target))
elif locked == False :
for i in range(int(popsize)):
generation.append(mutate(template, mutationrate))
for string in generation:
score = 0
for i in range(len(string)):
if string[i] == target[i]:
score = score + 1
else:
continue
stringscore = score, string
genscores.append(stringscore)
return sorted(genscores, reverse = True)
target = "Methinks it is like a weasel"
survivor = make_initial(target)
print "evolving ", target, " from ", survivor
generation = 0
fitness = 0
while fitness != len(target):
generation = generation + 1
nextgen = make_gen(survivor, target, 100, 0.1, True)
fitness, survivor = nextgen[0]
print "Generation %i: %s %i matches" % (fitness, survivor, generation)
print "Target evolved in", generation, "generations"
Labels: Dawkins, Evoution, mutation, python, sci-blogs, weasel


